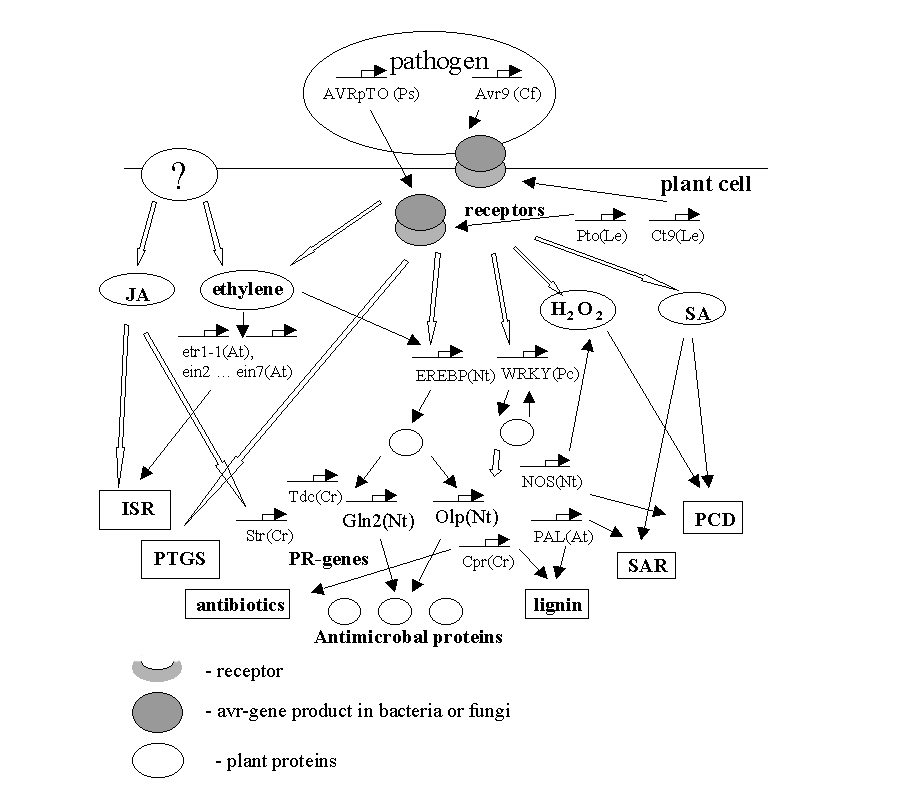
GENE NETWORK ON PLANT INTERACTION WITH PATHOGEN ORGANISMS
During the contact between plant and pathogenic microorganism, a particular chain of events is produced in the plant organism. The interaction between plant and pathogen may develop by two ways given below.
1. The plant is provided by a receptor that interacts with bacterial protein. As a result, quick protective reaction is being developed. In such a situation, the bacteria is called avirulent for a given plant genotype [Piffanelli P. et al., 1999, Martin G.B., 1999].
2. The proteins of the pathogenic organism are virulent for the given plant genotype. The plant is affected by the pathogen, whereas protective mechanisms are being activated more slowly [Maleck K and Lawton K., 1998].
In both cases, with the start of pathogenesis gene transcription, the cell walls strengthen. Then in the place of pathogen penetration, the active forms of oxygen are formed, causing the death of infected cells.
At the present moment, the best studied are the molecular mechanisms providing hypersensitive response (HR) [Halterman D.A. and Martin G.B., 1997]. In this case, the plant receptor interacts with the pathogen molecule. In order such interaction could occur, the plant and bacteria of a certain genotype should meet, i.e., a bacteria carrying the avirulence gene (avr) interacts with a plant, which has the corresponding R-gene. Such process is called an incompatible combination and leads to quick progressing of events, or to hypersensitive response. Receptors activate the passes of signal transduction and launch several protective systems. In the place of pathogen penetration, the strong oxidants are being synthesized such as H2O2, O2· - , OH· . Then the oxidative burst is being developed, followed by the death of infected cells according to the mechanism similar to apoptosis known in vertebrates [Lam E. et al., 1999]. Hypersensitive response is being developed in the place of pathogen penetration into the neighboring cells. Rapidly developing local process produces signal molecules spreading along vascular system of a plant.
The scheme of molecular events occurring during pathogenesis of plants. SA - salicylic acid; JA - jasmonic acid; H2O2 - reactive oxygen species; PR - pathogenesis-related genes; SAR -systemic acquired resistance; ISR - induced systemic resistance; PCD- programmed cell death; PTGC - post-transcriptional gene silencing. By thin arrows are given direct involvement of a substance of gene product in a reaction. Empty arrows denote the events that need additional elements for signal transduction.
 |
In dependence of local events, the set of signal molecules is being organized, which in turn, forms this or that generalized response. In the whole plant organism, the pathogenesis-related genes (PR-genes) are activated, the cell walls strengthen, and the plant accumulates some amount of protective substances, which are more effective in the struggle with this definite pathogenic form. In the plant cells, the salicylic acid (SA) is produced in considerable amounts and causes activation of SA-induced genes. The integrity of these events is named as systemic acquired resistance (SAR) [Mittler R. et al., 1999]. The other systemic, or referring to the whole organism, response to pathogen infection is an induced systemic resistance (ISR). Its differs from SAR by activation of some differing set of pathogenesis genes and by other ways of signal transduction (without participance of salicylic acid) [Pieterse C.M.J. et al., 1996].
In addition to immunity specific to pathogen, the general protective mechanisms are launched and, the plant becomes more resistant to other diseases after meeting the aviral pathogen [Zhu Q. et al., 1996].
The plant is not always supplied with receptors to the proteins of “attacking” bacteria or fungi. In this situation, the pathogen is called viral for given plant genotype; and the pair plant-pathogen is compatible. In this case, pathogen molecules are non-specific elicitors, which are non-specific substances causing pathogenesis. The ways of obtaining the signal from non-specific elicitors are still unknown. Various external stimuli (wound, non-specific elicitors) activate protein kinases and genes of signal molecules biosynthesis. In the course of signal transduction, the synthesis of JA, NO, H2O2, SA, and ethylene is produced. The processes are being activated that are well-known in animals: proteinkinase cascade, polyubiquitine-dependent protein degradation, etc. The signal transduction paths frequently intercross. For example, the gene of one of the key enzymes, PAL (phenylalanine ammonia-lyase), is activated not only in the course of hypersensitive response, but also in response to various external stimuli [Dixon R.A. and Paiva N.L. 1995, Mauch-Mani B. et al., 1996]. PAL takes part in the synthesis of SA, phytoalexins, and lignin monomers.
The latter, both in hypersensitive response and non-specific pathogenesis induction, activate pathogenesis-related (PR) genes. To these genes are referring those encoding enzymes of protective substances biosynthesis, chitinases acting in degradation of fungi cell wall, enzymes of lignin biosynthesis, which is a component of plant cell wall, etc.
The positive and negative feedbacks stabilize the parameters of gene networks. For example, WRKY, transcription factor proceeding in PR-genes activation is regulated according to the principle of positive feedback, because it has the binding site in its own promoter. Along with H2O2 and salicylic acid biosynthesis, the events are developing, which lead to the death of infected cells. The cell dies together with the source of signaling molecules. Hence, the gene network terminates its functioning by the mechanism of the negative feedback.