Proc. III Intern. Conference of intelligent systems for molecular biology, California, US, 1995, pp. 197-205.
Computer tool FUNSITE for analysis of eukaryotic regulatory genomic sequences
Kel A.E., Kondrakhin Y.V., Kolpakov F.A., Kel O.V., Romashenko A.G.,Wingender E.a), Milanesi L.b), Kolchanov N.A.
Institute of Cytology and Genetics, Siberian Branch, Russian Academy of Sciences, 630090 Novosibirsk, Russia, e.mail: kol@cgi.nsk.su, fax: (3832) 356558
a) Gesellschaft fur Biotechnologische Forschung mbH, Maschroder Weg 1., D-38124 Braunschweig, Germany, E.mail: ewi@venus.gbf-braunschweig.d400.de
b) Instituto di Tecnologie Biomediche Avanzate, CNR, via Ampere n.56, 20131 Milano, Italy
Abstract
We present the computer tool FunSite for description and analysis of regulatory sequences of eukaryotic genomes. The tool consists of the following main parts: 1) An integrated database for genomic regulatory sequences. The integrated database was designed on the basis of the databases TRANSFAC [1] and TRRD [2] that are currently under development. The following functions are performed: i) linkage to the EMBL database; ii) preparing samples of definite types of functional sites with their flanking sequences; iii) preparing samples of promoter sequences; iv) preparing samples of transcription factors classified with regard to structural and functional features of DNA binding and activating domains, functional families of the factors, their tissue specificity and other functional features; v) access to data on mutual disposition of cis-elements within the regulatory regions. 2) The second component of FunSite tool is the set of programs for analysis of the structural organization of regulatory sequences: i) Program for revealing of potential transcription factors binding sites based on their consensi; ii) program for revealing of the potential binding sites using homology search with nucleotide sequences of real binding sites; iii) program for analysis of oligonucleotide context features which are characteristic of flank sequences of the binding sites; iv) program for analysis of correlations between functional site positions; v) program for design of recognition method for the functional sites based on generalized weight matrix; vi) program for revealing potential composite elements. The results of analysis of the promoter sequences of eukaryotic genes with the FunSite are presented, too.
Introduction
Great effort has been made to unravel complete genomes, such as those of baker's yeast (Saccharomyces cerevisiae), mouse-ear cress (Arabidopsis thaliana), mouse or, with particular emphasis because of the medical impact, the human genome. The genome of Homo sapiens comprises approximately 3*109 base pairs. However, the pure sequence is of poor informational value if not accompanied with functional data. Thus, we have to know where a gene is located, between which positions its transcribed regions are placed, where its coding region starts, where it is interrupted by introns, where it terminates.
No due understanding of the structural organization of the genomes is possible unless we can recognize the regulatory regions that control gene transcription.
There is a definite methodological repertoire to reveal the structural features of transcription regulatory regions and the control mechanisms they are subject to. However, considering the large number of genes in the human genome, as well as the corresponding figures for the mouse and yeast and all other genomes, it becomes evident that we need efficient tools to deduce information about transcription regulatory regions from mere DNA sequences.
To our present knowledge, most of the control mechanisms of transcription regulation are mediated through a large variety of relatively short DNA sequence elements of 5 to 25 base pairs [1, 2]. Cluster of such modules constitute promoter or enhancer regions. These short regulatory sequences exert their effects (via events) at several distinct levels. At the first level, these elements are recognized by transcription factors whose sequence specificity is rather relaxed. The biological meaning of this degeneracy is to ensure a considerable variability in the efficacy by which distinct genes are expressed. This flexibility and, in some cases, even promiscuous ambiguity in the regulation mechanisms is required, e.g., for the tuned realization of the 105 genes of the human genome. Next, these factors act in a positive or negative manner onto the basal transcription initiation complex and, finally, on the activity of the RNA polymerase through a hierarchy of protein-protein interactions. A complex structure of the regions of gene transcription regulation arranges for this hierarchy. It is the structure of the regulatory regions that holds encoded all potentially possible ways of regulation of the gene under various conditions of expression (tissue specificity, stage of organism development, stage of cell cycle etc.)
The most important feature of organization of the transcription regulatory regions is their modular structure [3] and recognized levels of hierarchy. The lowest level in the hierarchy corresponds to cis-regulatory element, which binds a definite transcription factor. Really, most transcription factors bind to DNA as dimers which provides considerable variability of the interaction between transcription factors and their binding sites. For example, the bZIP class of trans-acting factors interact with the DNA through a positively charged (basic) region and, as a prerequisite, have to dimerize through hydrophobic interfaces called leucine zippers. One large subgroup of the bZIP factor family, the CREB - AP1 - factors, comprises at least 19 polypeptides that are able to form more than 37 dimers of different composition. The second level corresponds to composite response elements [4]. In most cases, the composite element is formed of adjacent or partially overlapping sites for proteins which belong to different factor families and to different signal transduction pathways. At the level of composite elements, a lot of ways for gene expression regulation are offered. Cross-coupling of two different factors at a composite element exhibits a new pattern of transcription regulation, for example, tissue-specificity of hormonal induction, tissue-specificity of immune or acute-phase response, etc. In some cases, one of the binding sites forming a composite element is low affinity binding site This is accounted for by stabilization of binding of the transcription factors to DNA because of additional protein-protein interactions, which opens more complex ways of gene regulation.
Several composite elements and/or individual binding sites are combined in promoters and enhancers. At the level of these structures, the specificity and multiple ways of regulation are provided by the entire set total of the sites and composite elements. Finally, the highest level of hierarchy is integrity of all the regulatory regions of the gene.
Thus, a huge variability of DNA-protein and protein-protein interactions provides a highly complex pattern for the regulation of gene expression in eukaryotic organisms.
The understanding of the transcription regulation mechanisms, so complex, and factors controlling transcription, so numerous, can be achieved via databases on transcription factors and transcription regulation regions.
Effective computer methods for analysis of this information are required, too. We have developed the computer tool FUNSITE which comprises two main component parts: 1) an integrated database for genomic regulatory sequences; 2) programs for analysis of the structural organization of regulatory sequences.
The integrated database on genomic regulatory sequences.
There is available nowadays a range of databases on genomic regulatory sequences: TRANSFAC [1], TRRD [5], TFD [2] and EPD [6]. The integrated database we report is based upon the first two.
The database TRANSFAC collects data on regulatory genomic sites and on transcription factors and thus consists of the two main records SITES and FACTORS, to which several additional records are connected. The additional features they contain are deduced from the basic mechanisms of gene regulation.
As data management system for the TRANSFAC data set, we have developed TRANSFAC retrieval program (TRP). It is a network-model system that ensures very rapid data access through physical links ("sets") between related entries. Many-to-many relations between records are established through common member records that may hold additional data qualifying the relation [7].
Transcription regulatory region database (TRRD) was developed to provide the comprehensive research on mechanisms controlling eukaryotic gene expression on the transcriptional level. In this database we collect data concerning various features of regulation of gene expression, gene classifications, structure of the gene regulatory regions, composite elements etc.
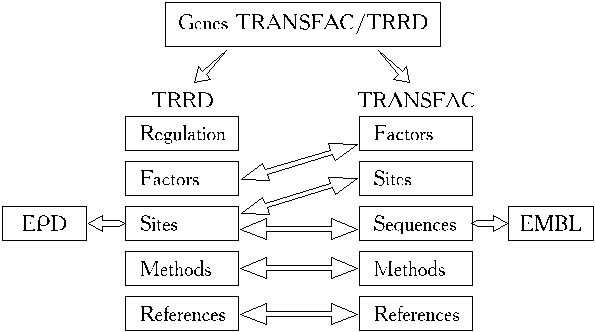
How the integration of TRANSFAC and TRRD was achieved is schematically presented in Fig.1. An additional table, GENES, contains the list of all the genes described in both databases. The GENES allowed one-to-one correspondence (Fig.1., double arrows) to be set up between the tables of TRANSFAC and TRRD.
The integrated database TRANSFAC/TRRD is linked to the EMBL data library and to the SWISSPROT database as well as to EPD. The eukaryotic promoter database (EPD) contains precise data on many eukaryotic promoters that have been experimentally revealed; it indicates transcription initiation sites and classification of all promoters described by functional and structural similarity and in accordance with some common features of gene regulation. Linkage between EPD, EMBL and TRANSFAC/TRRD is denoted in Fig.1 by single arrows. The following basic functions of the integrated database can be specified: i) linkage to the EMBL database; ii) preparing samples of definite types of functional sites with their flanking sequences; iii) preparing samples of promoter sequences; iv) preparing samples of transcription factors classified as regards structural and functional features of DNA binding and activating domains, functional families of the factors, their tissue specificity. and the functional features; v) access to data on mutual disposition of cis-elements within the regulatory regions.

Figure 1. TRANSFAC and TRRD integration scheme.
Computer programs for analysis of the genomic functional regions.
We have developed computer programs that have been included into the FUNSITE system. The system contains the following main programs for analysis of the characteristics of regulatory genomic regions. i) Program for revealing potential transcription factors binding sites based on their consensuses; ii) program for revealing the potential binding sites by using homology search with nucleotide sequences of real binding sites; iii) program for analysis of oligonucleotide context features which are characteristic of the sequences flanking the binding sites; iv) program for analysis of correlations of base frequencies at different positions of the functional site; v) program for construction of a recognition method for the functional sites based on a generalized weight matrix; 6) program for revealing potential composite elements.
Revealing potential transcription factor binding sites by consensus
To reveal potential transcription factor binding sites, we have used a compilation of transcription signals [8]. The compilation consists of k = 136 consensi of binding sites. The consensus lengths range from 5 bp to 24 bp, the consensi are described by using a 15-lettered code. The program developed detects all regions similar to the consensus in the nucleotide sequence under study. The program reveals potential binding sites that contain few nucleotides mismatching the consensus. The percentage of mismatches was designated as t. This parameter allows the functional variability of binding sites to be considered. By using t we can reveal poor binding sites not matching perfectly its consensus, yet are functionally active. On the other hand, the higher t, the higher the number of binding sites erroneously identified as being active, that is why the correct choice of t is of great importance here.
By using this method, we have analyzed a sample of polymerase II RNA promoters. A sample of P=470 promoters each of 600 bp in length from vertebrate genes was selected from EPD database. We selected sequences with lengths over 500 bp upstream of the transcription start and over 100 bp downstream of the transcription start. Then we break the promoters into subregions and analyze the concentration of transcription signals in each. The parameter m stands for the number of the promoter subregions. In our study m was ranging from 50 to 80. Analysis has revealed that a range of binding sites within the promoters are distributed unevenly. The unevenness was estimated by Smirnov's statistic W 2 [9]. The example of the uneven distribution of one well-known binding site Oct-1 in the promoter sample is presented in Figure 2. A large number of unevenly distributed potential transcription factor binding sites is characteristic of the eukaryotic promoter structure. Noteworthy, the binding sites for wide spread transcription factors AP2, TBP, Sp1, NF-IL6, E2A, EGR-1, HSF, NF-1, Oct-1 are found among the most unevenly distributed sites.
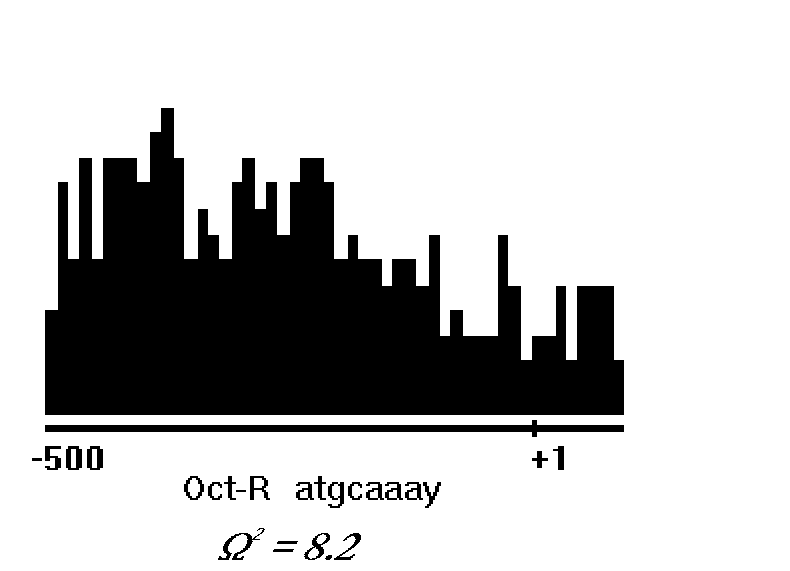
Figure 2. The example of the uneven distribution of binding sites for Octamer family members in the promoter sample
Recognition of potential transcription factor binding sites by patterns of real sites.
As a first step in the development of the method, we set up a compilation of aligned sets of patterns of real transcription factor binding sites. Some sets of the real sites from the compilation are presented in Figure 3. The recognition procedure was as follows. If a region of DNA in question exactly matches one of the sequences of the set of patterns, the region will be identified by the program as being a potential binding site. The compilation was built up as follows.
1) The sites that were experimentally shown to bind transcription factors of one subfamily were selected from the databases TFD, TRANSFAC and TRRD. By saying "a subfamily" we mean a group of closely related transcription factors with very similar binding sites in vertebrate species. We considered only those subfamilies for which more than 6 binding sites have been reported.
2) Find the core motif common to all sites of a given subfamily (see Fig.3: for the GATA subfamily, this motif is GAT trinucleotides)
3) Align all the sequences of the subfamily relative to the motif (without gaps).
4) To form a set of patterns from the sample of sites, we heed the following steps: (i) Extension of the current window w at either direction by one nucleotide from the core motif. (ii) Determine the most frequent sequence Sw within the current window W for the given sample of sites. (iii) The sequence fragments within the window will be considered as the current set of patterns. Sequences are included only for sites that have not more than 1 mismatch with the most frequent sequence Sw within the current window w. The other sequences of sites (if any) are omitted from the set of patterns. (iv) Check the first type errors of the recognition procedures. The first type error was evaluated on the basis of the number of sites omitted from the sets of patterns.
5) Iterations terminate and the ultimate set of patterns is thought to be built if the first type error is not over 10%.
GATA AP-2 ATF/CREB Sp1 agataag (4) ccccaggc (5) tgacgt (28) ccgccc (29) agatagc (2) cTccaggc (2) tgacgA ( 5) tgataag (2) cccGaggc (2) tgacgG ( 2) tgatagc (1) ccccTggc (2) tgacgC ( 2) tgatTag (1) ccccagCc (2) tgGcgt ( 1) tgataaA (1) ccGcaggc (1)
Fig.3. Examples for sets of patterns from the compillation. In parentheses: so-many sites of the given subfamily match the given pattern.
Comparative analysis of overprediction errors (second type errors) in the recognition of potential binding sites was performed for each of the subfamilies of transcriptional factors. To evaluate second type errors we used a representative sample of eukaryotic exon sequences of totale length 348000bp. The sample consisted of only internal exons of genes where no real transcriptional factor binding sites are practically observed. Table 1 presents first a 1 and second a 2 type errors for all the sites in question and, to compare with, the prediction errors for the sites by using the consensus method. As is seen, our method is advantageous over the consensus method. The point is that our approach takes into account the correlation between the positions within the sites. Really, as is seen for the compiled GATA-1 binding sites, the last two positions are described by RS (Fig. 3), but because of a correlation between nucleotides at the two positions, this description is not so detailed as the set of the real sites. In reality, only AG or GC occurs at these sites at the given positions, and none of AC or GG that fit in with the RS consensus. Thus, the overprediction error following the real site method is lower. However, the consensus method allows one to recognize potential binding sites for such factors that are not yet properly studied and for which just few real sites as these are known. The similar aproach for revealing of transcription factor binding sites was used in the work of Prestridge and Burks [10] where they also look at density of transcriptional elements in promoter and non-promoter regions.
Table 1. Examples of calculation of the first and second type errors at recognition by the consensus method and site pattern method.
Consensus method |
Site pattern method |
||||||
| Factor subfamily name | Consensus |
a1 *(%) |
a2 **(%) |
a1 |
a2 |
||
Exon set |
Promoter set |
Exon set |
Promoter set |
||||
| AP-2 ATF/CREB E2F GATA Sp1 |
cccmnsss tgacgyma ttttsscgs WGATAR KRGGCKRRK |
54.0 51.0 17.0 39.0 52.0 |
0.469 0.002 0.005 0.113 0.071 |
0.819 0.001 0.007 0.137 0.167 |
34.0 5.0 5.0 15.4 12.5 |
0.068 0.120 0.004 0.037 0.101 |
0.075 0.110 0.006 0.047 0.203 |
*
- the recognition error of the first type; ** - the recognition error of the second type
Recognition of Pol II promoters by using binding sites for transcription factors.
Compilation of transcription signals [8] which consists of 136 consensi of transcription factor binding sites is used for recognition of Pol II promoters. As was described above, we divide the promoter region into subregions and analyze the concentration of transcription signals in each of them. The parameter m that we used in our model is the number of analyzed subregion in the promoters. On the bases of the analyzed set of promoters we build the matrix:
T=|Tij|, t=|tij|; i=1,k; j=1,m.
where Tij is the number of incidences of the i-th signal (i = 1,136) in the j-th subregion through the all set of promoters.
Based on the matrix T, we have constructed a method for recognition of promoter regions. For each subsequence Z=(z1,z2,..., zL), where L=600 bp, we calculate a measure of it being similar to the promoter sample:
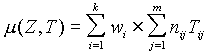 ,
,
where Tij are the elements of matrix T; nij is the number of occurrences of the i-th signal in the j-th region of the sequence Z; wi is the weight coefficient for the i-th signal. The weight wi is ascribed to the i-th transcription signal X=(x1,x2,...,xh) of length h, where xr is a symbol of a 15-letter code. The weight wi is calculated as follows:
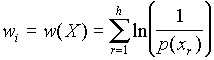 ,
,
where p(xr) is the probability of the letter xr occurring in a random sequence. We assume p(A)=p(T)=p(G)=p(C)=1/4; p(R)=p(Y)=p(W)=p(S)=p(M)=p(K)=1/2; p(B)=p(V)=p(H)= p(D) =3/4; p(N)=1. So, the signals that have a lower probability of occurrence in a random sequence get the higher weight w.
Let Z be a sequence under consideration. Let R1,R2,...,RP be a set of random sequences. The parameter m is evaluated for every random sequence r1=m (R1,T), ..., rP=m (RP,T) . The estimate b*=max{m (Ri,T)} is used as a threshold value of m for identification of Z. If m (Z,T) > b*, then sequence Z is identified as a promoter. For a more precise threshold value, we use random sequences simulated on the basis of the dinucleotide frequencies typical of the promoters of the sample. Random sequences generated in this way and the promoters are similar in contextual features, but the former are certainly unable to function as true promoters. Predicting ability surely rises, since we can now distinguish promoters from other sequences with very similar contextual characteristics.
Table 2 presents results of a check of the method. The first type error was estimated on the set of 470 promoters by using of the jack-knife method. The second type error was estimated on the sample of exon sequences which was descriped upper. As is seen, it provides higher accuracy than the method based on revealing of TATA and CAAT boxes [11]. Besides, we grouped the promoters with similar patterns of distribution of the signals along the sequences [12]. Eight groups were determined. The recognition method reinforced by this division exhibits a lower first type error and the same second type error.
Table 2. Testing of the methods for recognition of vertebrate promoters.
| Method | I type error (a 1) |
II type error (a 2) |
| Recognition by TATA box | 48% |
10.8% |
| Recognition on the basis of potential binding
sites without clusterization |
34% |
9.3% |
| Recognition on the basis of potential binding
sites after clusterization |
11%
|
9.5%
|
Analysis of the context features of sequences flanking transcription functional sites.
The system FUNSITE offers the following means to analyse the context features of functional sites with: 1) a program for analysis of the unevenness of the distribution of short oligonucleotides along the site; 2) a program for determination of dinucleotide weight consensi; 3) the program for revealing of oligonucleotides in 15-letter code typical of definite regions of sites.
As an example, we consider a sample of CRE elements from vertebrate genomes. The sample was created using the TRANSFAC database and EMBL data library. All the sequences were uniform in length 98bp, and contained the core sequence TGACgt at position 46. The sample consisted of 31 sequences. All the sequences in the sample are aligned to optimal matching to the TGAC core sequence.
Analysis of short oligonucleotide distribution
Smirnov's statistic W 2 mentioned above was applied to the analysis of the distribution of short oligonucleotides along the sequences containing CRE elements. A strongly nonrandom distribution of di- and trinucleotides along the sequences of CRE elements was revealed. As is seen from the Figure 4, the most typical features of the sequences flanking CRE elements are represented by the distribution of the short stretches CCC and TTT. The stretch CCC occurs in left-hand flanking sequences of the CRE element with abnormally high frequencies. The stretch TTT is rather confined to right-hand flanking sequences.
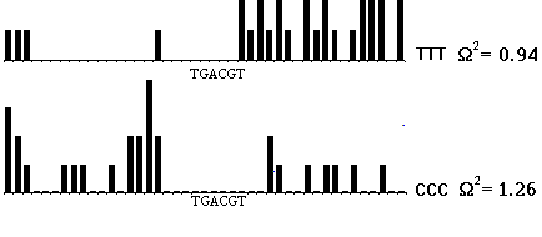
Fig. 4. The observed distribution of trinucleotides in CRE element flanking sequences. Present 43 positions around the core of CRE element. Each colon coresponds to one position. Height of the colones gives frequence of the given trinucleotide starting from the correspondent position.
Construction of dinucleotide weight consensuses.
A dinucleotide weight consensus is a vector ![]() , where
, where ![]() is the
weight attributed to the dinucleotides XY. The weights result from a training sample of
functional sites
is the
weight attributed to the dinucleotides XY. The weights result from a training sample of
functional sites  , where
, where ![]() is the number of the dinucleotides XY
occurring at position i over all sequences of the sites in question;
is the number of the dinucleotides XY
occurring at position i over all sequences of the sites in question; ![]() is the function of relative
"importance" of the positions, that defines the relative contribution of the
i-th position to the dinucleotide weight consensus. The unimodal functions
is the function of relative
"importance" of the positions, that defines the relative contribution of the
i-th position to the dinucleotide weight consensus. The unimodal functions ![]() were used in this analysis. While constructing
dinucleotide weight consensuses, the genetic algorithm is employed. With this algorithm,
we define the functions of position importance so that the resulting dinucleotide weight
consensus differs most from the distribution of dinucleotide frequencies in an alternative
sample. We use the exon sequences as the alternative sample in the analysis. Fig.5
presents bar diagrams for two dinucleotide consensuses, typical of the left-hand and
right-hand flanking sequences, respectively. The corresponding functions of
"importance" of positions are given there, too. As is seen from Fig.5, WW type
dinucleotides are largely typical of the left-hand flank of the site, while SS type
dinucleotides are typical of the right-hand flank.
were used in this analysis. While constructing
dinucleotide weight consensuses, the genetic algorithm is employed. With this algorithm,
we define the functions of position importance so that the resulting dinucleotide weight
consensus differs most from the distribution of dinucleotide frequencies in an alternative
sample. We use the exon sequences as the alternative sample in the analysis. Fig.5
presents bar diagrams for two dinucleotide consensuses, typical of the left-hand and
right-hand flanking sequences, respectively. The corresponding functions of
"importance" of positions are given there, too. As is seen from Fig.5, WW type
dinucleotides are largely typical of the left-hand flank of the site, while SS type
dinucleotides are typical of the right-hand flank.
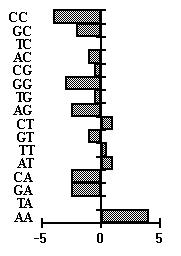
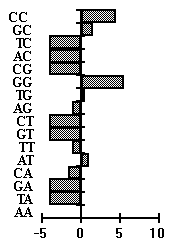
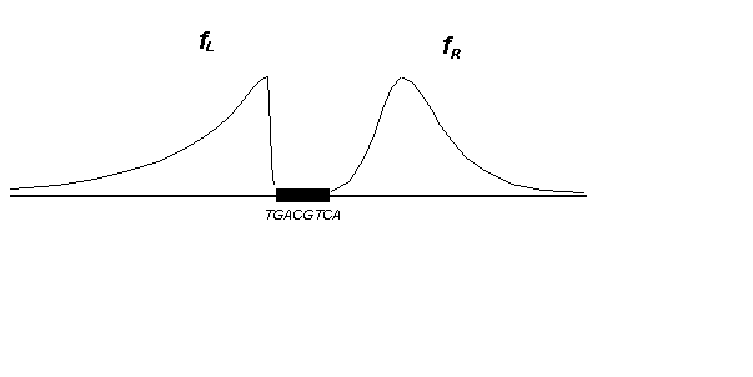
Fig. 5. Dinucleotide weight consensuses. a) CRE element left -hand flank; b) CRE element right -hand flank. fL , fR -function of position "importance" for the left-hand and right-hand flank.
Revealing oligonucleotides in a 15-letter code
Analysis of the set of CRE elements was performed by using the SITEVIDEO program [13] which detects oligonucleotides in 15 letter code typical of definite regions of CRE elements. We found that the oligonucleotide C G/T A -T (CKAV) is less frequent in the region around the CREB binding site (Fig.6). Thus, regions about 15bp around the CRE-element is significantly purred from this pattern. We believe there is strong selection against this particular signal to prevent binding of some transcription factor which prevents CREB-transcription factor from being bound
On the other hand, we have found that the two triplets, CGA and CTA, that form part of the revealed oligonucleotide CKAV, occur at very low frequency in the set of known transcriptional cis-elements. The promoter region around the CRE element probably has a very specific oligonucleotide composition that lowers chances of random incidence of any binding site in the course of the mutational or recombinational process. Such "clearing" of the region around CRE elements could be of importance for the precise positioning of transcription factors binding to the site.
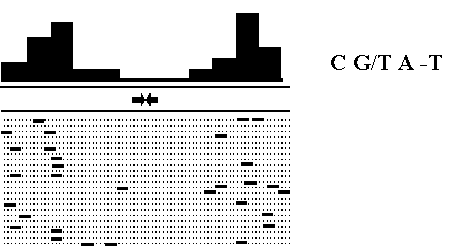
Fig.6 Concentration of CKAV oligonucleotide around CRE-element (in the center).
Recognition of transciption regulation sites with the matrix of a generalized consensus.
This method is an extension of the well-known method of site
recognition on the basis of the weight matrix [14]. Consider this method as applied to the
recognition of CRE elements. Training involves a sample of sites, each being 98 bp in
length. Let Z={AA, AT, ..., CG, CC} be the set of 16
all possible dinucleotides (d=2). Then ![]() ,
, ![]() is the matrix of the generalized consensus, where the element
is the matrix of the generalized consensus, where the element ![]() is the frequency of the incidence of a H-type
oligonucleotide (HÎ Z) at position j of the sample of functional sites.
Regarding the task, this matrix of oligonucleotide frequencies is obviously better than
the commonly accepted matrix of mononucleotide frequencies in that it takes into account
the correlation between neighbouring nucleotides. Each row defines the distribution of a
definite dinucleotide along the site. First, the profile of the oligonucleotide
distribution is subject to smoothing. The smoothing procedure involves the parameter u
which sets the size of the smoothing window. The other two parameters of the method are w
which stands for the length of the fragment under study and s which stands for the
localization of the core sequences within these fragments. For the nucleotide fragment X
of length w, its similarity with the CRE elements is defined by the following
multiplicative measure
is the frequency of the incidence of a H-type
oligonucleotide (HÎ Z) at position j of the sample of functional sites.
Regarding the task, this matrix of oligonucleotide frequencies is obviously better than
the commonly accepted matrix of mononucleotide frequencies in that it takes into account
the correlation between neighbouring nucleotides. Each row defines the distribution of a
definite dinucleotide along the site. First, the profile of the oligonucleotide
distribution is subject to smoothing. The smoothing procedure involves the parameter u
which sets the size of the smoothing window. The other two parameters of the method are w
which stands for the length of the fragment under study and s which stands for the
localization of the core sequences within these fragments. For the nucleotide fragment X
of length w, its similarity with the CRE elements is defined by the following
multiplicative measure
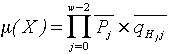 ,
,
where ![]() , if
, if ![]() ; and
; and ![]() , if
, if ![]() .
. ![]() , if
, if ![]() ;
and
;
and ![]() , otherwise. Here Hj is the type of dinucleotide in the j-th position of the
testing sequence X.
, otherwise. Here Hj is the type of dinucleotide in the j-th position of the
testing sequence X.
This measure states to what degree the fragment under study is similar
with the set of functional sites as regards oligonucleotide distributions. If ![]() , then the fragment may be
regarded as a CRE element. Table 3 presents the results of testing the method reported
under various parameter values.The first type error a 1 was estimated by the jack-knife
method. The second type error was estimated on two control samples. One consisted of the
exon sequences from vertebrate genes (error a 21). The other sample consisted of the same
exon sequences with the core sequence of CRE element inserted (error a 22). In this way,
we generated false CRE elements that matched the consensus but did not perform the
function of the true CRE elements.
, then the fragment may be
regarded as a CRE element. Table 3 presents the results of testing the method reported
under various parameter values.The first type error a 1 was estimated by the jack-knife
method. The second type error was estimated on two control samples. One consisted of the
exon sequences from vertebrate genes (error a 21). The other sample consisted of the same
exon sequences with the core sequence of CRE element inserted (error a 22). In this way,
we generated false CRE elements that matched the consensus but did not perform the
function of the true CRE elements.
Table 3. Testing the method of recognition of CRE elements with the matrix of the generalized consensus.
Recognition errors |
||
Parameters |
a 1(%) |
a 21(%) a 22(%) |
u=3, w=30, s=9 |
25.0 |
0.43 38.0 |
The results obtained by this method demonstrate that it provides some advantages if compared with the consensus method or site pattern method. It can be used as an additional test for the signals revealed with consensuses or real sites.In fact, the generalised consensus method effectively disregards false signals found in exons (with an error of 38 % only), whereas the other methods erroneously identify these signals as being CRE elements.
A program for recognition of potential composite elements.
Recognition of pair-wise combinations of binding sites appears to promise more accurate recognition. Functional sinergism between the closely located sites and the cooperative binding of factors to these sites had been experimentally shown in many cases. Experimental data demonstrate that the distance between two binding sites within the composite elements does not exceed 50-60bp, and is much less in most cases. That is why the recongnition of potential composite elements is based upon the search of promoters for the wide-spread pairs of binding sites in which the sites are not more than 50 bp apart.
The sites were sought for throughout the promoter sample by the site pattern method. The promoter sample consisted of N = 470 sequences of length L=600 bp each. The following procedure was applied to each pair of the sites S1 and S2. A window of w=50bp was moving along all the promotor sequences of the sample. At any situation of the window, the sites S1 and S2 were sought for in the sequence within the window. Then we performed an analysis of the correlated incidence of the two signals so near, for which we counted the number of detections of these signals in four possible situations.
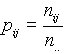 , i,j =0,1
, i,j =0,1
where n.. = N(L - w +1) is the number of all situations of the
window over all promoters in which the sites S1 and S2 were looked for; ![]() is the number of the situations of the window
when none of the signals was present;
is the number of the situations of the window
when none of the signals was present; ![]() - is
the number of the situations of the window when S1 is missing and S2 is
present;
- is
the number of the situations of the window when S1 is missing and S2 is
present; ![]() - is the number of the situations
of the window when S1 is present and S2 is missing;
- is the number of the situations
of the window when S1 is present and S2 is missing; ![]() - is the number of the situations of the window when both signals
are present.
- is the number of the situations of the window when both signals
are present.
On the basis of the frequencies, statistic c 2 is calculated
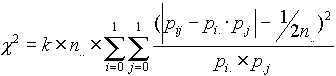 ,
,
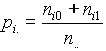 ,
, 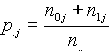 , i,j = 0,1
, i,j = 0,1
k is the coefficient of renormalizing which is used to remove the
effect of multiple account of data which takes place as the window moves along the
sequence ![]() . If
. If ![]() , where
, where ![]() is the threschould value, we
acknowledge a correlated incidence of the two binding sites in the promoters at a distance
of less than 50bp. The most interesting pairs are those for which the observed frequencies
of concurrent incidence in the promoters is significantly higher than expected (
is the threschould value, we
acknowledge a correlated incidence of the two binding sites in the promoters at a distance
of less than 50bp. The most interesting pairs are those for which the observed frequencies
of concurrent incidence in the promoters is significantly higher than expected (![]() ). These pairs were called
potential composite elements. Table 4 presents examples of the elements we have found. As
is seen from the Table, some of the pairs correspond to the experimentally detected type
of composite elements. Besides, for some pairs we did not find experimentally evidence in
literature, but these pairs may perform certain functions. A number of experimentally
detected composite elements is present in TRRD database.
). These pairs were called
potential composite elements. Table 4 presents examples of the elements we have found. As
is seen from the Table, some of the pairs correspond to the experimentally detected type
of composite elements. Besides, for some pairs we did not find experimentally evidence in
literature, but these pairs may perform certain functions. A number of experimentally
detected composite elements is present in TRRD database.
Table 4 Examples of potential composite elements.
Potential composite element |
|
|
Experimentally detected |
|
S1 |
S2 |
|||
| AP-1 AP-2 HNF1 HNF1 ATF/CREB ATF/CREB ATF/CREB C/EBP GATA IRF-1 Oct |
GR Sp1 GR NFIII E4F1 ETFA Sp1 Sp1 NF-kB NF-kB Sp1 |
24.95 36.41 88.01 37.99 33.79 11.50 30.83 8.32 20.01 33.46 5.36 |
1.205 1.999 1.340 1.672 9.987 3.044 2.525 5.590 5.712 18.082 4.080 |
+ a)
+
+
+ + |
a) + - this pairs correspond to the experimentely detected type of composite elements
Figure 7 presents the localizations of the potential composite elements formed by AP-1 and RAR binding sites. This type of composite elements was found in promoters of different genes. As is seen from the Figure 7, the distance between sites is quite fixed and small, which again provides evidence that it is a functionally coupled signals. In most cases, the potential composite elements revealed are located between -200 and +100 relatively the transcription start site. This region must be corresponding to the most functionally active region of the gene promoter.
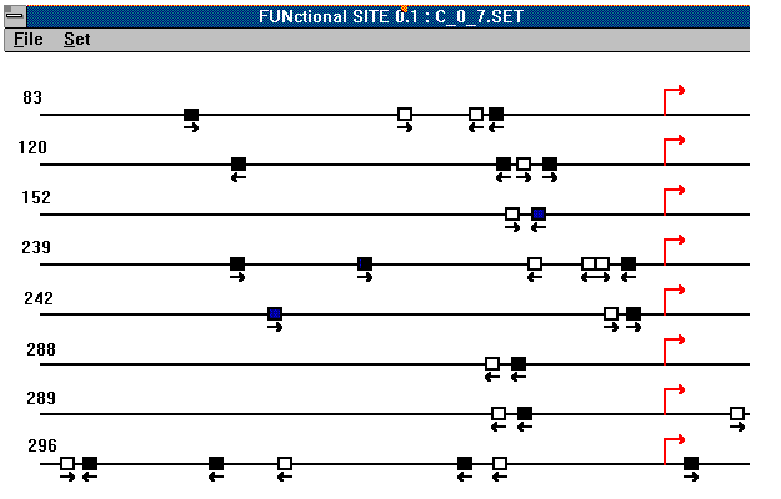
Figure 7. Localization of the potential composite elements formed by AP-1(white) and
RAR (black) binding sites in promoter sequences (from -500 to +100). Statistical
significance: ![]() Arrows indicate transcription
start points. Genes are: 83 - human islet amiloid polipeptide gene; 120 - human gene for
epsilon-globin ; 152 - mouse polimerase beta beta gene; 239 - rat alpha amilase (AMY-1)
gene; 242 - rat neuron-specific anolase gene 288 - bovine luteinizing hormone-beta subunit
gene; 289 - pig luteinizing hormone-beta subunit gene; 296 - ovine growth hormohe gene.
Arrows indicate transcription
start points. Genes are: 83 - human islet amiloid polipeptide gene; 120 - human gene for
epsilon-globin ; 152 - mouse polimerase beta beta gene; 239 - rat alpha amilase (AMY-1)
gene; 242 - rat neuron-specific anolase gene 288 - bovine luteinizing hormone-beta subunit
gene; 289 - pig luteinizing hormone-beta subunit gene; 296 - ovine growth hormohe gene.
Acknowledgements
This work was supported by the Russian Foundation of Fundamental Investigations (grant N 94-04-13241-a), Russian National Human Genome Project, Russian Ministry of Sciences and Tehnique Politics, USA Department of Energy, "Genetic Engineering Project" CNR, Italy, German Ministry of Sciences.
References
1. E.Wingender Recognition of regualtory regions in genomic sequences. (1994) Journal of Biotechnology, v. 35, p. 273-280.
2. Gosh D. Status of the transcription factors database (TFD) (1993) Nucl. Acids Res., v.21, n.13, p.3117-3118.
3. Dynan W.S., Modularity in promoters and enhancers ( 1989) Cell, v.58, p.1-4.
4. Diamond M.I., Miner J.N., Yoshinaga S.K., and Yamamoto K.R. Transcription factor interactions: selectors of positive or negative regulation form a single DNA element. (1990) Science, v.249,p.1266-1272.
5. Kel O.V., Romachenko A.G., Kel A.E., Naumochkin A.N., Kolcanov N.A. Data representation in the TRRD - a database of transcription regulatory regions of the eukaryotic genomes. Proceedings of the 28th Annual Hawaii International Conference on System Scienses [HICSS]. (1995), v.5, Biotechnology Computing, IEE Computer Society Press, Los Alamitos, California, p.42 -51
6.Bucher,P. EPD: Eukaryotic promoter database. (1993) Current release.
7. Knueppel et al., (1994)
8. Faisst,S., Meyer, S. Compilation of vertebrate-encoded transcription factors. (1992) Nucl. Acids Res., v. 20, p. 3-26.
9. Darling D.A. (1957) Ann.Math.Statist., v. 28, p.823-838.
10. Prestridge and Burks (1993), Hum. Mol. Genet., v. 2, p.1449-1453
11.Bucher,P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. (1990) J.Mol. Biol., v. 212, p.563-578.
12. Y.V.Kondrakhin , A.E.Kel , N.A.Kolchanov, A.G.Romashchenko, L.Milanesi Eukaryotic promoter recognition by binding sites for transcription factors. (1993) CABIOS, in press.
13. Kel A.E., Ponomarenko M.P., Likhachev E., Orlov Yu.L., Ischenko I.V., Milanesi L., Kolchanov N.A. SITEVIDEO: a computer system for functional site analysis and recognition. Investigation of the human splice sites (1993) CABIOS, v. 9, n. 6, p. 617-627.
14.Shapiro, M.B. and Senapathy P. RNA splice junctions of different classes of eukaryotes:sequence statistics and functional implications in gene expression. (1987) Nucleic Acids Res,. v.15, p. 7155 - 7174.